本招聘信息由金沙8888js官方2008级博士王利民提供
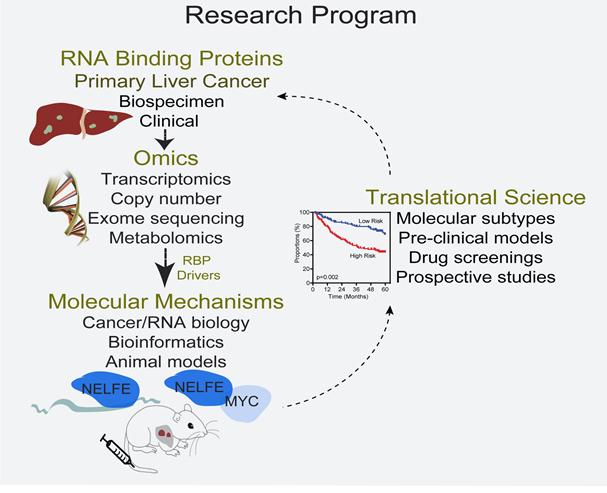
Summary: Transcriptomic imbalance is pervasive in cancer. An integral group of proteins that regulate the transcriptome is RNA binding proteins (RBPs). Wehave previously demonstrated that manyRBPs are dysregulated in a subset of primary liver cancers (PLCs) with poor prognosis, suggesting thatRBPs are selected during tumor evolution.PLC is the 2nd most common cause of cancer-related death worldwide with the fastest rising incidence and mortality in the United States. Despite considerable efforts towards improving diagnosis and developing new treatment modalities, PLC remains among the most difficult-to-treat malignancies, with a 5-year survival rate of less than 15%. Our lab is interested in identifying disease-acquired functions of RBPs, and in parallel, exploit such mechanisms as actionable biomarkers or drug targets to improve diagnosis for PLC patients.Our lab utilizes bioinformatics and bench science spanning from functional genomics, translational science, molecular epidemiology, RNA biology, cancer biology, and animal models to investigate the oncogenic roles of RBPs in PLC (see Figure). Our main focuses are as follows:
Project 1:central focus of our research is investigating the dysregulation of mRNA binding proteins (mRBPs) in hepatocellular carcinoma (HCC). Our recent work identified Negative Elongation Factor E as an oncogenic RBP that is associated with poor clinical outcome in more than 1200 clinical specimens. We identified the novel mechanisms in which NELFE enhances the oncogene MYC to promote tumorigenesis. We are currently interested in how NELFE interact and regulate their RNA targets through modular domains such as low complexity sequences (LCS) and RNA recognition motifs (RRMs).
Project 2:Tumor heterogeneity represents a significant barrier to improving HCC patient outcome and poses a challenge for the establishment of robust HCC classification, making treatment extremely difficult. Consequently, the ability to discriminate patients with greater homogeneity and clinically relevant therapeutic targets will help guide treatments to improve patient outcome. We have developed a robust 20-NELFE dependent MYC target (NDMT) gene signature, which reflects the biological characteristics of HCC. Moreover, this signature has been validated in four independent cohorts consisting of more than 850 HCC patients with different etiological factors including race and ethnicity and showed a stronger prognostic value for survival in HCC patients than current clinicopathological prognostic factors. We are interested in utilizing high-throughput screenings to identify therapeutics for this specific subtype. In collaboration with others, we have developed a MYC-induced NELFE HCC Sleeping Beauty mouse model (Trp53flox/flox; Alb-Cre) that can be used to validate drug targets in vivo and patient derived cell lines that can used for screening.
Project 3:In addition, we are interested in utilizing several areas of RNA biology, translational, bioinformatics, and molecular biology to identify key oncogenic RBPs involved in transcriptomic alterations associated with disease states. For example, we have recently identified Argonaute 2 (AGO2) as a possible oncogenic RBP that can promote an HCC transcriptome in a RISC-independent manner. By utilizing transcriptomics, we show that activated AGO2 preferentially affects mRNAs by enhancing their stability. This is only evident in HCC with high AGO2 HCCs. We’re now elucidating how AGO2 promotes HCC progression by directly interacting with mRNA targets independent of miRNAs.
Postdoctoral Position
@ Thomas Jefferson University, Philadelphia, PA
新开实验室,发文章有保证。Dr. Hien Dang这个人也很厉害,目前已发表的一作Cancer Cell. 2017 Jul 10;32(1):101-114.e8,还有几篇在投的。她就坐在我旁边的,人非常好相处的,听说武大的学生都很优秀就想招。实验室的氛围特别好,学生之间关系也很好。她老板也是肝癌领域的大牛,在NCI领导美国liver cancer moonshot program。实验室详情请见课题介绍,欢迎有感兴趣的同学申请。由于目前的工作(NCI)将于8月底结束,而新单位(TJU)10月份报到,故请发送简历至htdangster@gmail.com
[打 印] [关 闭]
|